Vöhringer Lab
26 - November - 2021
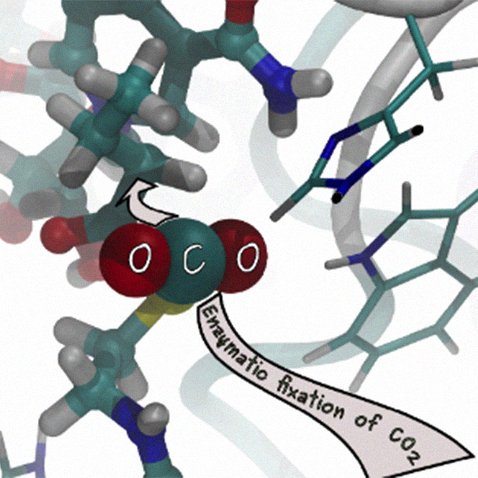
Enzyme Catalysis for efficient CO2 fixation
Carbon dioxide is one of the most abundant greenhouse gases associated with global warming. One of the main goals to reduce global warming’s adversary effects is to reduce its emission and develop alternative methods to transform this gas into valuable compounds.
In our lab we try to understand how enzymes fix CO2 and transform it to organic compounds with high efficiency. We have studied the reaction mechanism of RuBisCO one of the most abundant protein that fixes most of atmospheric CO2 in the Calvin cycle. Although abundant, RuBisCo is not the most efficient one. To learn from the best in Nature, we focused on the family of Enoyl-crotonyl-CoA carboxylase/reductase which possess the fastest fixation rate observed in enzymes and no side reaction with oxygen. At the end, we would like to disclose the whole catalytic cycle: possible conformational changes of the protein in the catalytic cycle, CO2 binding in the active site and the reaction mechanism to form the products.
To address the CO2 distribution in enzymes and the associated conformational changes, we work in a close collaboration with Prof. Grubmüller at Max-Planck Institute Göttingen in Germany in the established Computational Reaction Dynamics Max-Planck Partner group.
The reaction mechanism is elucidated from a computational point of view by us. Our results together with the biochemical characterization of the protein structure with Prof. Soichi in Stanford and the kinetics in the group of Prof. Erb at the Max-Planck Institute Marburg provide a detailed picture how these molecular machines are able to transform CO2 efficiently.
Our final goal is to understand how these enzymes work and then design new variants with enhanced efficiency to be applied in biocatalysis.
Improving force field description of molecular interactions for accurate free energy prediction
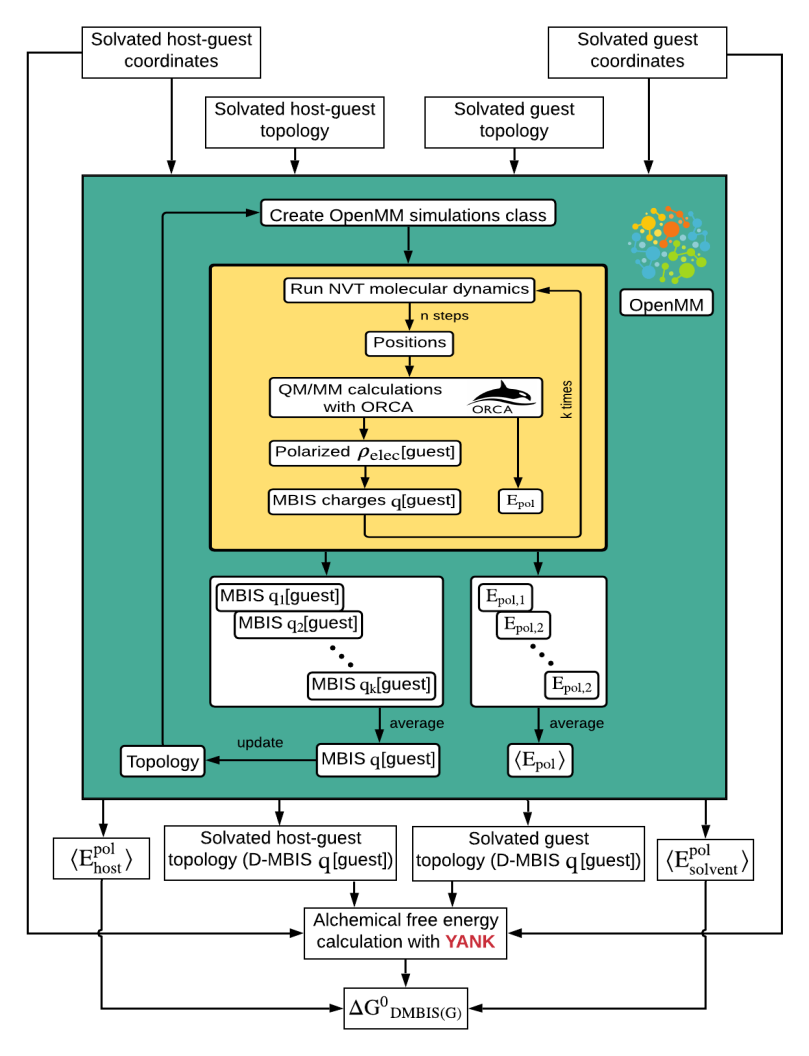
 In another proyect we develop new force fields for drugs and biolomolecular systems, which can be used to study their binding affinity or kinetics. We use electronic structure calculations and the Atom-In-Molecules approach to obtain force field parameters as atomic charges in close collaboration with Prof. Verstraelen in Ghent, Belgium, Dr. Fanaz Heidar-Zadeh in Queens University, Canada and Dr. Paul Ayers in Mcmaster University, Canada in the QC-devs community.
In another proyect we develop new force fields for drugs and biolomolecular systems, which can be used to study their binding affinity or kinetics. We use electronic structure calculations and the Atom-In-Molecules approach to obtain force field parameters as atomic charges in close collaboration with Prof. Verstraelen in Ghent, Belgium, Dr. Fanaz Heidar-Zadeh in Queens University, Canada and Dr. Paul Ayers in Mcmaster University, Canada in the QC-devs community.
As a first step we focused on hydration free energies and partition coefficients in a close collaboration with Dr. David Mobley at UC Irvine. We could show that our approach provides parameters, which reproduce hydration free energies and partition coefficients as good as standard parameters in established force fields.
Recently we have also addressed the CB7 and octa acid host-guest systems. Our non-bonded force field parameters are able to improve the absolute binding free energy prediction of several guest to the host (see publications).